Pheatmap in R: Create Customizable Clustered Heatmaps
Updated on
Heatmaps are an essential tool in the data scientist's toolkit, providing a visually intuitive representation of complex datasets. Among the various packages available in R for generating heatmaps, Pheatmap stands out for its flexibility and customization options. This article will guide you through the process of creating beautiful, customizable clustered heatmaps using Pheatmap in R.
Pheatmap is more than just a function in R; it's a powerful tool that allows users to create clustered heatmaps with greater control and customization options than the standard R heatmap function. With Pheatmap, users can visualize gene expression analysis, draw correlation heatmaps, and customize label sizes and dendrogram visibility. Let's dive into the world of Pheatmap and explore its capabilities.
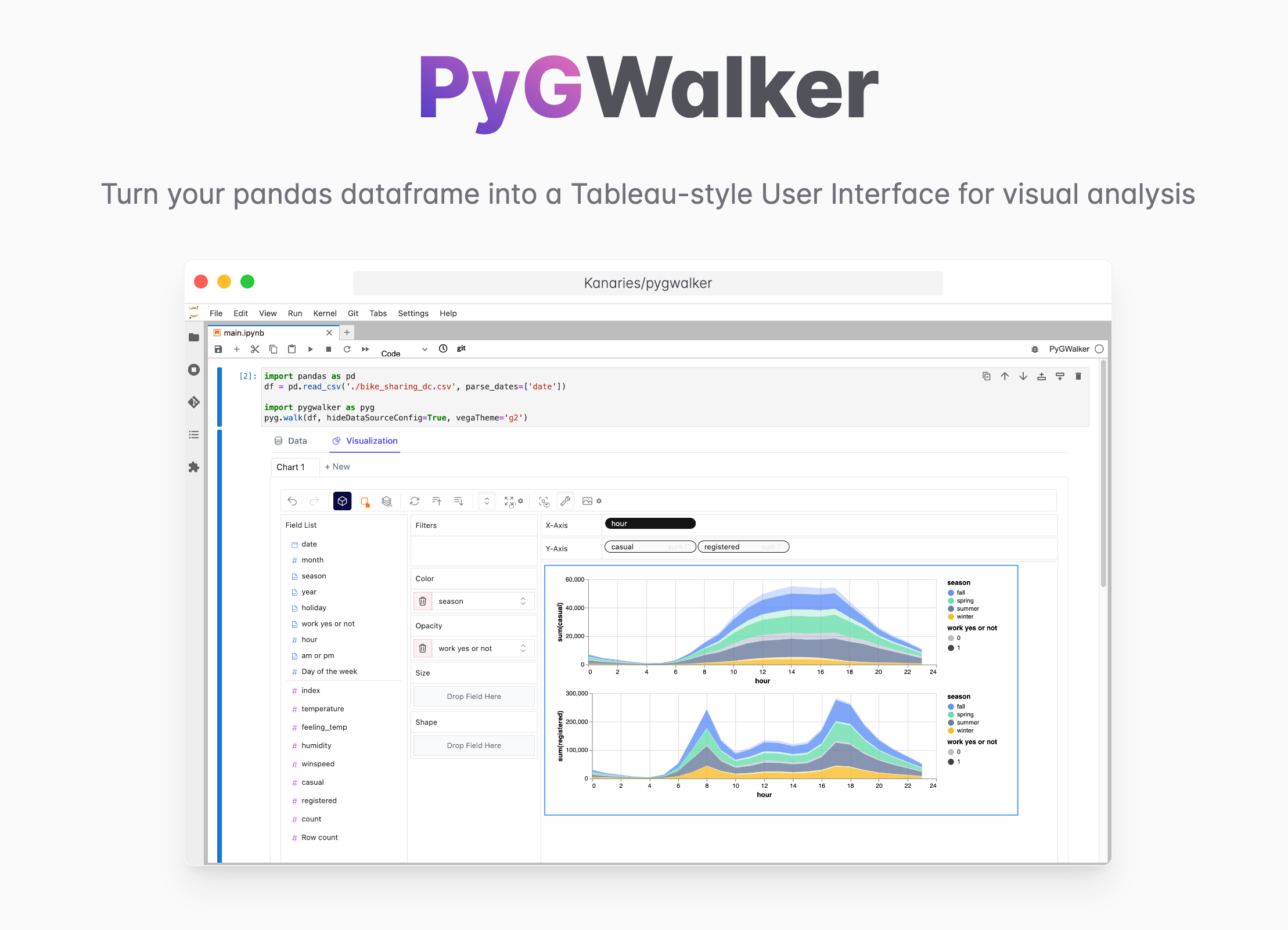
Want to quickly create Data Visualization from Python Pandas Dataframe with No code?
PyGWalker is a Python library for Exploratory Data Analysis with Visualization. PyGWalker (opens in a new tab) can simplify your Jupyter Notebook data analysis and data visualization workflow, by turning your pandas dataframe (and polars dataframe) into a tableau-alternative User Interface for visual exploration.
What is Pheatmap in R?
Pheatmap is a function in R that generates pretty heatmaps, allowing data scientists to visualize complex data in a simplified manner. It offers more control and customization options compared to the standard base R heatmap functions like heatmap() and heatmap.2(). Pheatmap stands out for its ability to produce aesthetically pleasing and informative heatmaps.
Pheatmap is particularly useful in genomics, where it is often used to visualize gene expression data. It allows for the addition of annotations and uses clustering methods to group similar data, enhancing the interpretability of the heatmap. It also provides options for row/column Z-score standardization, which can be crucial in certain data analysis scenarios.
How does Pheatmap work?
Pheatmap works by taking a matrix of data and converting it into a visually intuitive heatmap. The data values are represented as colors in the heatmap, with the color intensity indicating the magnitude of the value. This allows for easy identification of patterns and correlations in the data.
The function also performs hierarchical clustering on the data, grouping similar rows and columns together. This is represented visually by a dendrogram, a tree-like diagram that shows the hierarchical relationship between the data points. The clustering method used by Pheatmap can be customized according to the user's needs.
Pheatmap also allows for a high degree of customization of the heatmap's appearance. Users can control the color palette, label sizes, dendrogram visibility, and more. This makes Pheatmap a versatile tool for data visualization in R.
Advantages of Pheatmap over standard R heatmap
While the base R heatmap function is useful for basic heatmap generation, Pheatmap offers several advantages that make it a preferred choice for many data scientists.
Firstly, Pheatmap provides more control over the appearance of the heatmap. Users can customize the color palette, adjust label sizes, and control the visibility of the dendrogram. This allows for the creation of heatmaps that are not only informative but also visually appealing.
Secondly, Pheatmap performs hierarchical clustering on the data, grouping similar rows and columns together. This enhances the interpretability of the heatmap and allows for easier identification of patterns in the data.
Thirdly, Pheatmap allows for the addition of annotations and the use of filters, which can be particularly useful in gene expression analysis. It also provides options for row/column Z-score standardization, offering more flexibility in data analysis.
In conclusion, while the base R heatmap function is a useful tool for basic heatmap generation, Pheatmap offers a
higher level of control and customization that makes it a powerful tool for data visualization in R.
Customizing the Appearance of Pheatmap in R
One of the key advantages of Pheatmap is the ability to customize the appearance of the heatmap to suit your specific needs. Here's how you can do it:
Color Customization
Pheatmap allows you to customize the color palette used in the heatmap. This can be done using the color parameter in the pheatmap() function. You can choose from a variety of color palettes available in R, or create your own.
Label Customization
The size and appearance of labels in the heatmap can be adjusted using the fontsize and fontface parameters. This allows you to control the readability of the heatmap and adjust it according to your presentation needs.
Dendrogram Visibility
Pheatmap allows you to control the visibility of the dendrogram, a tree-like diagram that shows the hierarchical relationship between the data points. This can be done using the show_rownames and show_colnames parameters in the pheatmap() function.
Adding Annotations
Pheatmap allows you to add annotations to the heatmap, which can be particularly useful in gene expression analysis. This can be done using the annotation_row and annotation_col parameters in the pheatmap() function.
In conclusion, Pheatmap provides a high level of customization that allows you to create heatmaps that are not only informative but also visually appealing. Whether you're visualizing genomic data or drawing correlation heatmaps, Pheatmap offers the flexibility and control you need to create beautiful, customizable clustered heatmaps in R.
Clustering Method Used by Pheatmap
Pheatmap uses hierarchical clustering to group similar data points together. This is a method of cluster analysis that seeks to build a hierarchy of clusters. The end result is a tree-based representation of the data, called a dendrogram, which allows users to visualize the data in a way that highlights the relationships between the data points.
In Pheatmap, the clustering method can be customized using the clustering_distance_rows and clustering_distance_cols parameters for rows and columns respectively. The default method is "euclidean", but other methods like "maximum", "manhattan", "canberra", "binary" or "minkowski" can also be used.
Plotting Heatmaps in R with Pheatmap
Creating a heatmap with Pheatmap in R is straightforward. Here's a basic example:
# Load the pheatmap library
library(pheatmap)
# Create a matrix of data
data <- matrix(rnorm(200), 20, 10)
# Generate the heatmap
pheatmap(data)This will generate a basic heatmap with default settings. You can customize the heatmap by adding parameters to the pheatmap() function. For example, to change the color palette, you can use the color parameter:
# Define a color palette
my_palette <- colorRampPalette(c("blue", "white", "red"))(25)
# Generate the heatmap with the custom color palette
pheatmap(data, color = my_palette)Customizing Colors in Pheatmap
Pheatmap allows for a high degree of color customization. You can define your own color palette and apply it to the heatmap. This is done using the color parameter in the pheatmap() function. Here's an example:
# Define a color palette
my_palette <- colorRampPalette(c("blue", "white", "red"))(25)
# Generate the heatmap with the custom color palette
pheatmap(data, color = my_palette)In this example, the colorRampPalette() function is used to create a palette of 25 colors ranging from blue to white to red. This palette is then applied to the heatmap using the color parameter.
Related guides
- Clean and reshape data before plotting: R dplyr Data Wrangling Pipeline
- Choose the right chart type fast: R ggplot2 Quickstart
- Import CSV, Excel, or Parquet with correct types: Importing Data in R
Conclusion
In conclusion, Pheatmap is a powerful tool for creating customizable clustered heatmaps in R. Whether you're visualizing genomic data, drawing correlation heatmaps, or just exploring your data, Pheatmap offers the flexibility and control you need.
Frequently Asked Questions
What is the advantage of using Pheatmap over the standard base R heatmap?
Pheatmap offers several advantages over the standard base R heatmap function. It provides more control over the appearance of the heatmap, performs hierarchical clustering on the data, and allows for the addition of annotations and the use of filters. This makes it a powerful tool for data visualization in R.
How can I customize the color palette in Pheatmap?
You can customize the color palette in Pheatmap using the color parameter in the pheatmap() function. You can choose from a variety of color palettes available in R, or create your own.
What clustering methods does Pheatmap use?
Pheatmap uses hierarchical clustering to group similar data points together. The clustering method can be customized using the clustering_distance_rows and clustering_distance_cols parameters. The default method is "euclidean", but other methods like "maximum", "manhattan", "canberra", "binary" or "minkowski" can also be used.